Just because scientists know the gene responsible for a rare disease doesn’t mean they know what’s going on. NGLY1 is a good example. A handful of children around the world have an inherited deficiency in NGLY1, leading to a complex neurodevelopmental disorder. A family diagnosed at Emory thought their older affected daughter had cerebral palsy for most of her childhood. How the loss of an enzyme that removes sugar tags from certain proteins causes problems is still being uncovered.
More broadly, geneticists can read a family’s DNA sequences and find the differences, but figuring out what they mean is still a challenge. That’s where the approach taken in a recent paper in Cell Systems, by Emory cell biologist Victor Faundez and colleagues at Illinois State, comes in.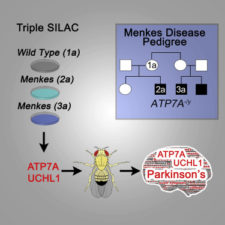
His lab used a comparison of proteomes to dissect Menkes disease, a rare inherited deficiency in a copper transport enzyme called ATP7A. This means they didn’t compare genes; instead, they compared the proteins produced by patients’ cells with those in their unaffected relatives.
Copper is an essential part of the diet and can be found at the active sites of several enzymes. The symptoms of Menkes disease arise because of a lack of copper in the body, although cultured cells lacking ATP7A actually accumulate copper.
The Cell Systems paper shows an overlap between the protein abnormalities coming from copper accumulation and cellular defects in some forms of familial Parkinson’s disease. Both appear to impact mitochondria, the authors write.
The authors also caution that a single patient pedigree may not be sufficient to uncover disease mechanisms, unless the data set is expanded by increasing the number of affected families or the protein data is sifted by other means.
As Anette Breindl of BioWorld notes, “very rare diseases are hard to study in humans, because treatment groups are so small that within-group variability is likely to drown out any between-group differences.”
Breindl also notes: Faundez’s team intends to test whether their method can be used to gain insights into disorders that involve more than one gene, such as microdeletion syndromes. They also plan to compare the results they have obtained in fibroblasts to results in iPSC-derived neurons.

